Layout
The FragView layout consists of several toolbox and dropdown options to render and fragment molecules. The following image depicts the possible toolbox options that allow for erasing bonds, fragment a molecule, undo, redo, and 2D center buttons.
- Eraser: Allows for bonds connecting atoms in the 2D model to be deleted, which can then be set as a child node by clicking the Frag button. Note, the eraser does not allow for the deletion of atoms, only bonds.
- Frag: To create a new child node from the current node selected in the tree view on the right of the screen, press the Frag button. Any changes made to the molecule in the 2D view will be saved in the newly created node.
- Undo: To undo a change made to the 2D structure, including deleting a bond or loading another 2D structure by clicking a node on the tree, press button pointing to the left.
- Redo: To reverse an undo, the redo button allows for the 2D view of the model to be returned to a more recent state.
- Center: If the molecule ever becomes improperly centered in the 2D view, the center button allows for the focus to be brought to the molecule correctly.
Additional buttons and drop-down options can be found in the image below with their purposes described.

- Smile bar: The textbox shown on the far left of the image above is where the smile or smiles to be created will be entered. A new tree with one root node is created by specifying a single smile, while multiple children nodes can be generated along with the root by separating the smiles with periods.
- New: The new button will create a new tree using the smile or smiles in the smile bar. Note that the current tree will be replaced by the new tree, so if you want to save the data, make sure to go to Tools > Save tree data.
- Refresh: The refresh button will retake the current screenshot of the 2D view of the molecule and replace the node image in the tree view with the newly taken image. If the tree image is ever distorted, the node image can be recreated using this button.
- Tools: Various options for saving files related to the molecule.
- Model: Options that will change how the models are shown to the user.
- Help: Includes the link to the tutorial video and about page.
- The tree view on the right side of the applications will show a 2D image for the molecule of that node and allow the user to quickly navigate to parent or children nodes created. If multiple children nodes are created simultaneously in the smile bar using periods to separate children nodes from the root node, the root node cannot be loaded. To properly navigate to this node, the full root node must be created alone and then the fragments made individually. The currently active node will be represented by a purple box and clicking on another node will load the 2D and 3D models into their respective views.
Creating Trees
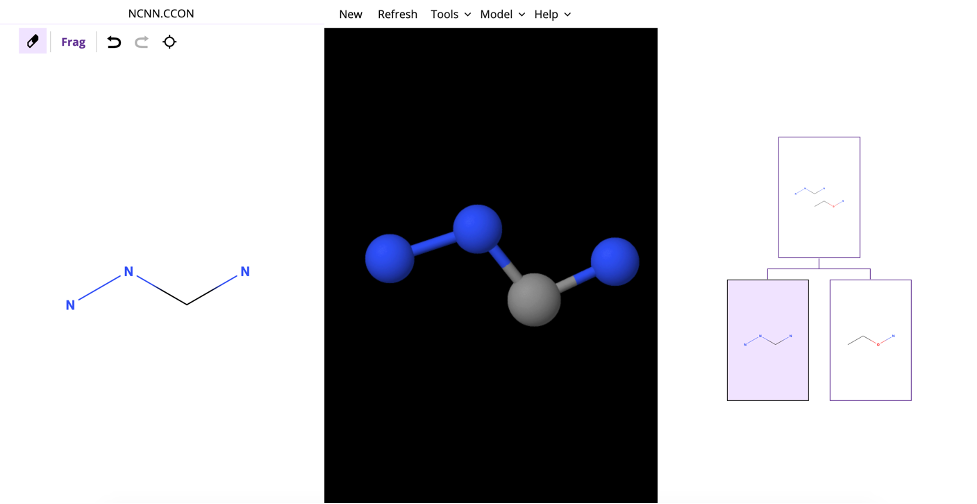
- FragView allows for trees to be created in two ways, including typing a smile or InChI into the smile bar, textbox in the top left corner of the application, or loading a previously saved tree. RDKit is used for the generation of molecule files, which means the program does not rely on a database service, such as Pubchem. Any smile can be entered into the textbox to create a new tree with the root node having a molecule with that smile. Furthermore, the tree can automatically be created with children by delimiting multiple smiles with periods. For instance, the NCNNCCON molecule might be the root node but typing NCNN.CCON in the smile/InChI bar will have a root node of NCNNCCON with two children molecules of NCNN and CCON. After the smile has been entered in the smile bar, press the new button to replace the current tree with the new tree specified by the smile or smiles in the textbox.
- To save a tree structure to work on it later go to Tools > Save tree data.
- Once you have the file saved, it can be uploaded to create a new tree using the Tools > Upload Tree Data feature.
Valence Electrons
- The Fragview 3D viewer keeps track of valence electrons that are added on from the original molecule when a new fragment is created. These electrons are colored yellow and stay yellow as more fragments are generated.
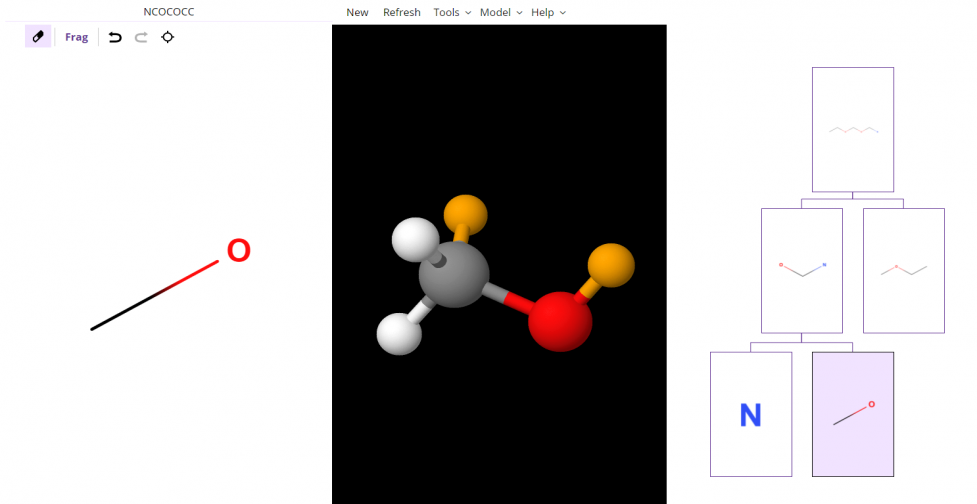
- In the following image, you can see where one valence electron was added to the NCO child after splitting the NCOCOCC molecule indicated by the color yellow.
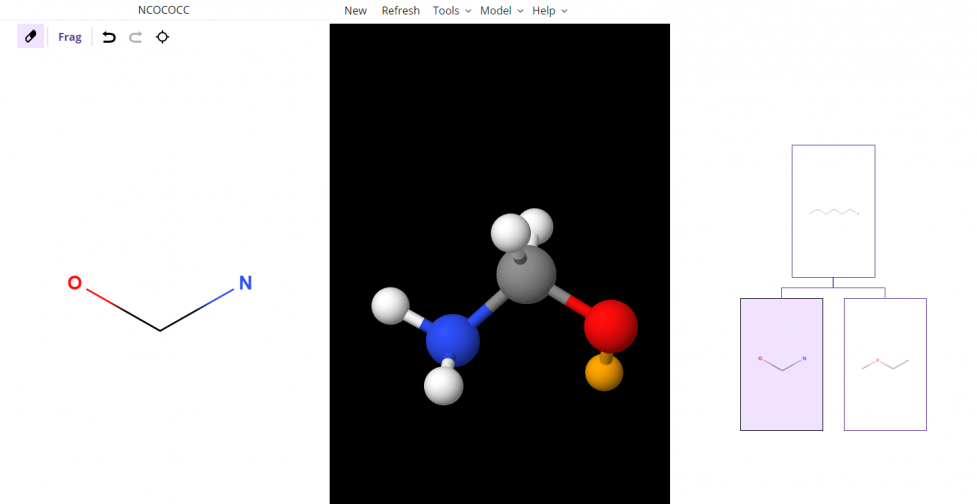
- The following image shows that if the NCO child is split again, the valence electron connected to an Oxygen atom from the previous example remains yellow. There is also a new valence electron added onto the Carbon atom.
Import/Export Options
Underneath the Tools menu, there are some options for importing and exporting data in Fragview.
- Structural formula image: PNG of Sketcher view.
- 3D model image: PNG of 3D viewer.
- MOL file: MOL file exported from RDKit.
- STL file: STL file of the 3D model of the molecule currently being rendered. Some more information can be found later in the manual.
- Save tree data: JSON file that stores entire tree in order to be imported back into Fragview.
- Upload tree data: Upload JSON file that restores a tree previously built in Fragview.
3D Viewer Settings
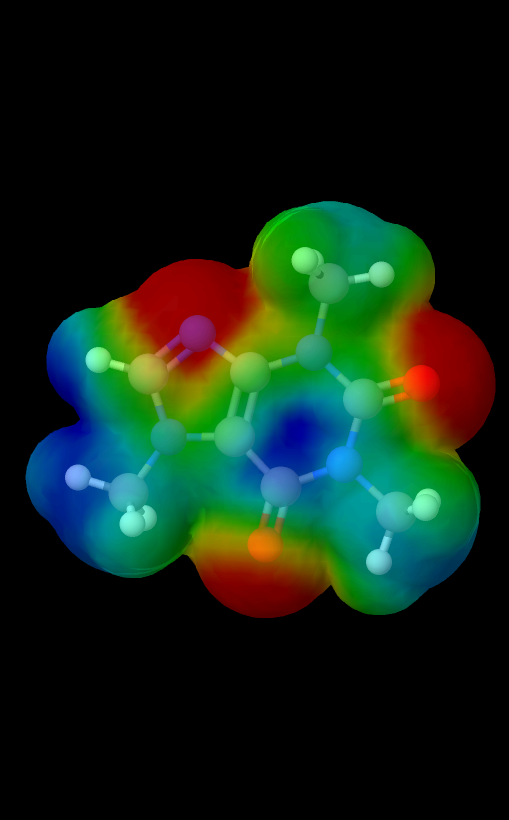
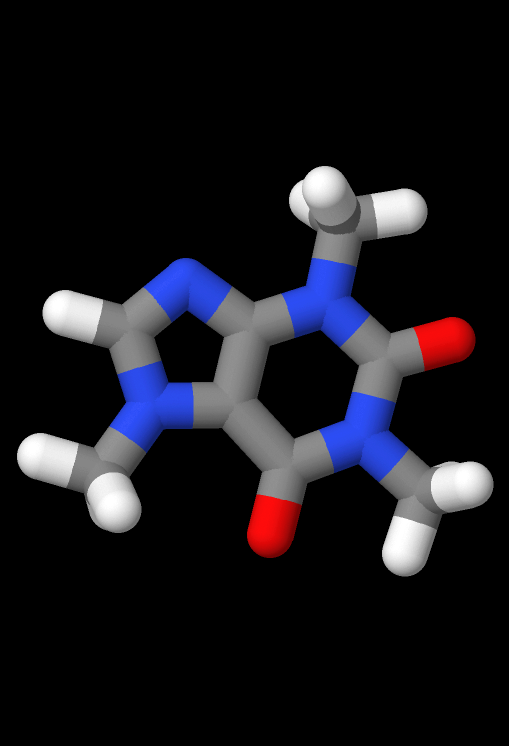
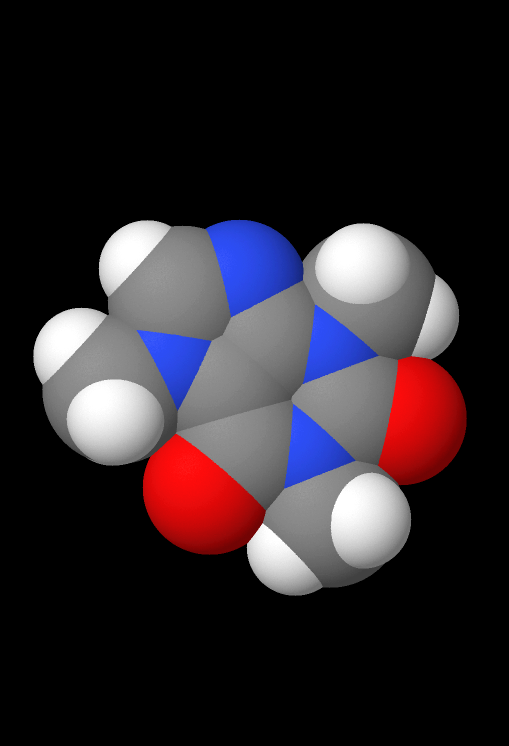
Underneath the Model option in the top menu, there are a few options that change the 3D viewing environement. All examples pictured below are visualizations of the caffeine molecule.
- Isosurface vdw: Toggles isosurface Van der Waals spheres visualization.
- Ball and Stick: Toggles the default ball and stick visualization.
- Stick: Toggles the stick visualization where the bonds and atoms are uniform in size.
- Van der Waals spheres: Toggles the Van der Waals spheres visualization.
- Wireframe: Toggles the wireframe visualization.
- Line: Toggles the line visualization that has no distinction between atoms and bonds.
- Background: There are three background colors available for the 3D viewer: Black(default), Gray, and White.
STL Printing
- FragView is able to generate an STL file for the currently showing molecule in the 3D viewer by navigating to the Tools > STL File for 3D Printing option. From there, a splicer software can be used to generate the gcode file and 3D print the molecule. The following image shows an example of the STL file retrieved from FragView.